CRISPR/Cas9 Editing Efficiency Increased by 60%

Researchers from UC Berkeley have improved CRISPR/Cas9 gene editing performance by 60%. The team designed a specific DNA template that has the desired sequence and additional nucleotides complementary to the DNA end that remains free after Cas9 cuts the double helix. The presence of the DNA template right at the editing site dramatically increases the chances of successful DNA substitution. THe study has been published in the journal Nature Biotechnology.
CRISPR/Cas9 is a gene-editing technology based on Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR), a prokaryotic immune system that cuts fragments of alien DNA and incorporates them in CRISPR loci. CRISPR transcripts (crRNA) bind and guide Cas endonucleases to cut the complementary DNA site. Currently, CRISPR/Cas9 is probably the most popular biolmolecular technique. However, its efficiency when incorporating DNA is much lower than for just cleaving it. Given the importance of base-pair mutations in genetic diseases, it would be very helpful to find a way of improving CRISPR’s performance when substituting DNA sequences.
Bringing the template DNA to the cut site
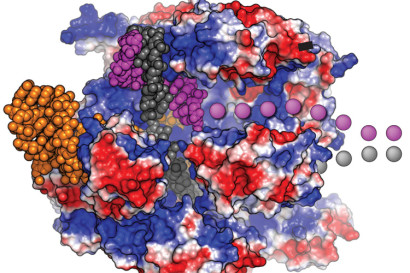
Christopher Richardson et al. found that Cas9 (in the picture, in red and blue) stays attached to the DNA (grey and purple) up to six hours after cutting it: it binds three DNA ends, leaving one free (purple dots). The Berkeley team designed a DNA template with the sequence of interest followed by some bases complementary to the free DNA strand. DNA was then recruited more efficiently to the site to be edited, increasing the number of effective substitutions. The team accomplished to edit a mutation with a 60% higher efficacy.
Finally, the researchers showed how Cas9 variants that don’t act as endonucleases can bind DNA and attract a template sequence. Using these Cas9 variants has the advantage of totally avoiding off-target DNA cuts.
Source: Berkeley