Feature Rich Tool Launched for Binder Design at Generative Speed – BoltzGen
Simulating molecular motion at atomic resolution has long been a cornerstone of computational chemistry, but it’s notoriously slow. Traditional molecular dynamics (MD) simulations, which numerically integrate Newton’s equations of motion, can take weeks or months to explore just nanoseconds of real molecular behavior.
From DiffDock to BoltzGen: The Next Leap in Generative Molecular Design
In 2023, MIT researchers introduced DiffDock, a powerful tool for performing molecular docking — fitting small molecules (potential drugs) into protein binding sites using a diffusion-based generative model. DiffDock was a landmark for computational drug design, accelerating virtual screening and reshaping how scientists predict drug–target interactions, however its success rate at binding was limited in many ways.
Researchers from same group have unveiled BoltzGen — a bigger, broader, and more capable successor. Where DiffDock focused on small molecule–protein docking, BoltzGen extends generative modeling to multiple molecular classes: not just small molecules, but also nanobodies, proteins, and peptides.
This expansion transforms BoltzGen from a docking tool into a universal generative framework for molecular structure and binder design, capable of producing equilibrium conformations and realistic complexes across molecular modalities.
Functionalized protein binders have emerged as versatile tools that can be used to target and manipulate proteins. Such protein binders can be based on various scaffolds, such as nanobodies, designed ankyrin repeat proteins (DARPins) and monobodies, and can be used to block or perturb protein function in living cells.
Before models like DiffDock and BoltzGen, designing molecular binders—such as antibodies, peptides, or small molecules that attach to specific biological targets—was a laborious process rooted in experimental screening and physics-based simulations. Researchers relied heavily on molecular docking algorithms, molecular dynamics (MD) simulations, and energy minimization to predict how a molecule might interact with its target.
These methods often required weeks of compute time and produced limited accuracy due to approximations in force fields and solvent effects. Experimental validation typically involved high-throughput screening of thousands of candidate molecules or directed evolution in the lab to improve affinity and specificity. In contrast, modern generative AI models like DiffDock and BoltzGen learn from structural data to predict or design binders directly, bypassing much of the computational and experimental trial-and-error.
A New Paradigm in Molecular Simulation and Design
Developed by researchers from the Massachusetts Institute of Technology (MIT) and Boltz Inc., BoltzGen represents a new generation of deep learning models that learn both molecular physics and binder design directly from data.
The model approximates the statistical mechanics of molecular structures and binding events by learning the Boltzmann distribution of 3D molecular configurations through deep generative diffusion modeling.
Once trained, BoltzGen can generate equilibrium molecular conformations and design functional binders in seconds — from nanobodies and cyclic peptides to small molecules and disulfide-bonded peptides — while maintaining rotational, translational, and permutational invariance.
Unprecedented Experimental Diversity and Performance
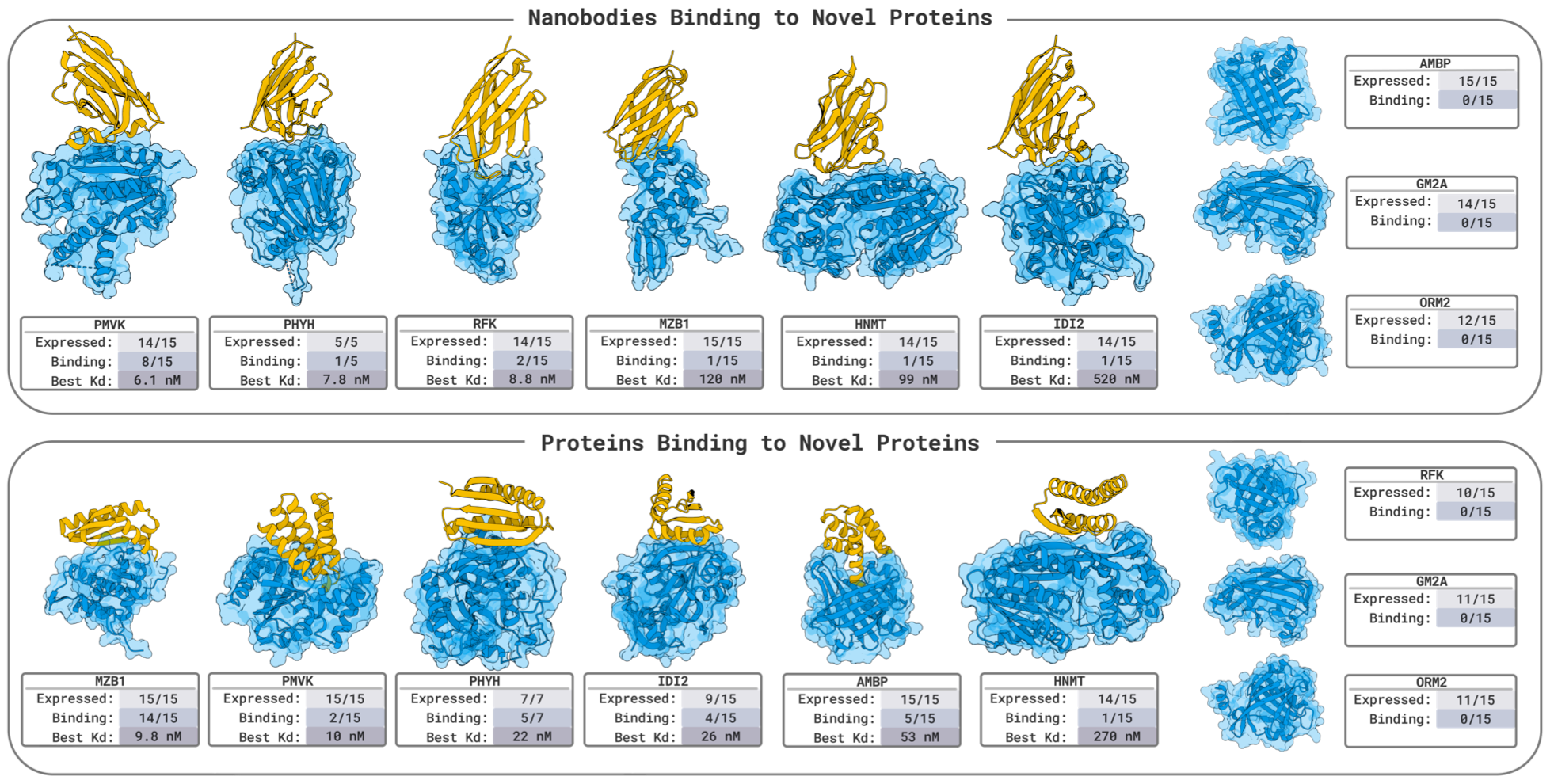
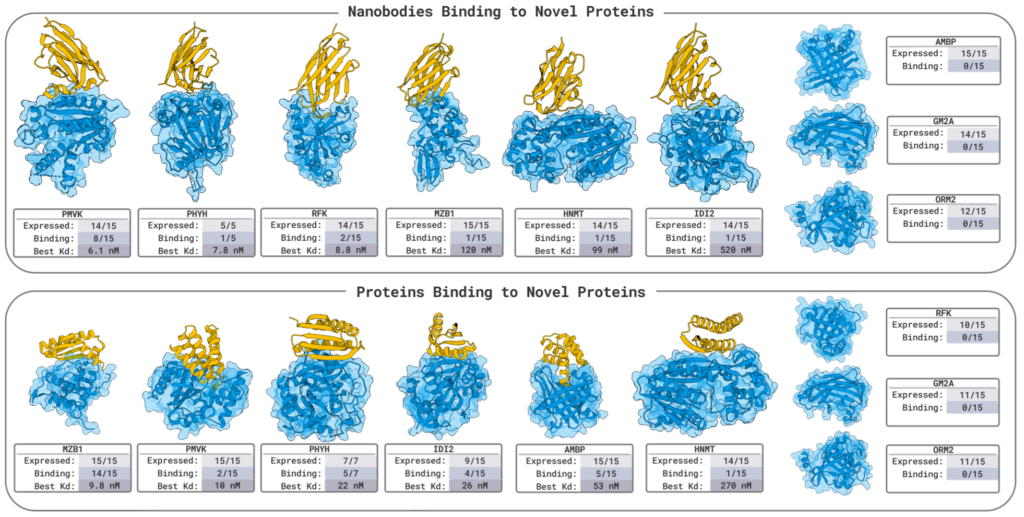
The breadth of BoltzGen’s experimental validation is unmatched. Its experiments span diverse binder modalities — from nanobodies to disulfide-bonded peptides — and include targets ranging from intrinsically disordered proteins to small molecules.
In benchmark tests, BoltzGen achieved state-of-the-art results, with a 67% success rate in designing nanomolar nanobody binders against several novel targets, each requiring fewer than 15 designs.
This marks one of the highest success rates ever reported for generative binder design and represents a leap from theoretical modeling to experimentally validated generative biophysics — including functional readouts in live cells.
Design Control Meets Generative Physics
Beyond speed and accuracy, BoltzGen introduces a design specification language that lets users guide the generation process using physical and structural constraints — such as bonding patterns, binding site geometry, secondary structure motifs, and design masks.
This makes BoltzGen not just a model for discovery but a programmable design engine — enabling users to generate molecules that satisfy both energetic plausibility and functional intent.
Its open-source release under an MIT License, with model weights, code, and data freely available, further cements BoltzGen’s role as a foundation for next-generation molecular design platforms. (boltz.bio).
How BoltzGen Works
BoltzGen in its current version supports four different default protocols:
protein-anythingfor designing proteins to bind protein/peptide targetspeptide-anythingfor designing peptides to bind protein targetsprotein-small_moleculefor designing proteins to bind small moleculesnanobody-anythingfor designing single-domain antibodies or “nanobodies”
These all protocols designs binders by proceeding through seven discrete steps:
- Design. Design initial binder structures according to input specification using a diffusion model (using the core BoltzGen model).
- Inverse folding. Predict sequences of amino acids that will fold into those structures (“inverse folding” using the BoltzIF model).
- Design folding. Refold the newly-predicted amino-acid sequences with their targets using Boltz-2 to validate that they’re actually predicted to bind the target.
- Folding. Refold standalone structures of newly-predicted amino-acid sequences (skipped if using the
peptide-anythingornanobody-anythingprotocols) to validate that the designed proteins will be stable on their own. - Affinity. Predict protein–ligand binding affinities (if using the
protein-small_moleculeprotocol). - Analysis. Analyze to predict design quality.
- Filtering. Filter and rank designs to select best candidate binders.
The Bigger Picture: Generative Physics Meets Molecular Design
BoltzGen’s architecture is dual-purpose, which signals a new era in computational science and joins the growing breed of Generative models and tooks that are shortening R&D times. It unifies structure prediction with structure design in one model.
By learning both the energy landscapes and binding principles that govern molecular behaviour, BoltzGen transcends the limitations of simulation-based methods. It delivers instantaneous access to thermodynamically valid ensembles and experimentally verified binder candidates, turning what once took months of MD and wet-lab iteration into a matter of hours.
Following in the footsteps of DiffDock, BoltzGen represents the next evolutionary stage of AI-driven molecular modeling — from docking small molecules to designing biologics and macromolecular therapeutics.
As generative models like BoltzGen, DiffDock, and GFlowNet continue advancing, they are blurring the line between simulation and synthesis, heralding a future where molecular discovery begins with generative physics.

