Open Microscopy App: Unifying Biological Image Data

High resolution images are ubiquitous in biological research, making the process of analyzing, saving, sharing and publishing them pivotal to research. Images promote scientific integrity, prevent duplication of effort and they help to disseminate scientific discovery. One organization is leading the fight to simplify biological image management and has been instrumental in creating a single, standardized access point for more than 130 image types. OME or Open Microscopy Environment provides an image format translation library called Bio-Format and a software platform for easy visualization called OMERO. Thousands of labs around the world employ OME’s programs for data access and management, making it an essential tool for every researcher. Following are the details about possibly the only tool that researchers will need to know about when it comes to handling biological images.
Bio-Format is an open-source Java library that allows almost any image processing software to read and edit biological file formats by allowing interoperability. It is probably the most comprehensive image translation library that supports over 130 image file formats and it is freely available to everyone. Helen Flynn, the OME Technical Writer describes it as a bridge between the acquisitions software and the software tools that a user might want to use to visualize or analyze their image data with. It ensures that you and others have access to your image data, even after the proprietary software packages no longer regularly update for latest operating systems and devices, by way of standardization.
The diverse software packages that support it include scripting software such as i3dcore, Qu for MATLAB and ITK; numerical data processing software such as KNIME, IDL, and MATLAB; visualization and analysis software such as Adobe Photoshop, Imago, Endrov, CellProfiler, FocalPoint, VisBio, XuvTools, and Vaa3D; and image servers such as its own OMERO and BISQUE. The complete list of software and file formats supported can be found here. It supports data from Brightfield and fluorescence microscopy, 3D cell division (color & time-lapse), multiple parametric analysis, digital pathology, high content screening and many more in its list of analysis techniques.
Bio-Format is supported by a very active open source community and commercial partners such as Glencoe Software. Installation is a simple two step process: downloading the .jar files from the OME website and pasting them in the software’s plugin directory. Experienced users are allowed to make changes by forking the project from its open GIT repository.
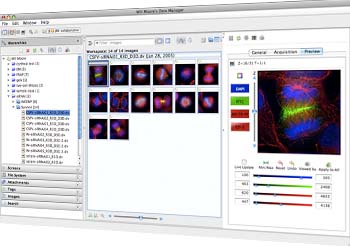
OMERO works in combination with Bio-Format, allowing users to access, analyze, organize, annotate, visualize, share and publish data. It allows the user to access data from a local file or remote servers. It is basically a middleware that runs in environments such as Java, Java Client, MATLAB and Python. It consists of a server, a web based client and server desktop clients for Windows, Linux, Mac OS X. Commercial patrons of OMERO include PerkinElmer, Rockefeller University Press, Applied Precision, American Society for Cell Biology and many others.
Update: Previously the GIT repository was wrongly linked to Gelncore’s open source files, it has now been corrected and is linked to OME’s own GIT repository.