LiteFold: Democratizing AI-Driven Protein Research

LiteFold is a unique AI-native research platform built not by traditional academic researchers, but by curious developers who personally encountered the operational friction of existing computational biology tools—fragmented workflows, minimal user interfaces, and poorly maintained documentation. What began as a seemingly interesting challenge has evolved into a platform that translates state-of-the-art methods in protein structure prediction, molecular design, and in-silico drug discovery into a streamlined, click-driven experience—combining intuitive, well-engineered user interfaces with scalable, production-grade cloud infrastructure.
While open-source protein structure and molecular design prediction models are widely available, running these models remains a major challenge for scientists, students, and innovators outside elite computational labs. Most of these tools lack a usable UI, require PhD-level biological knowledge, and demand expert-level software engineering skills to deploy and maintain. At its core, LiteFold represents a growing movement in biotech: shifting from expensive, fragmented pipelines to integrated, AI-powered research infrastructure that accelerates discovery while lowering technical barriers.
UI as a Strategic Differentiator and Complexity Reduction Layer
While tools for structure prediction and drug discovery are readily available, a recurring theme across LiteFold’s blog is reduced friction—not scientific friction, but operational friction:
- GPU setup: Not required, as LiteFold utilizes cloud-based GPUs
- Format incompatibilities: AI-assisted input fields help users understand errors and correct them in real time
- Toolchain hopping: Move from literature review to protein design to drug discovery within a single platform
- Long feedback cycles: Real-time progress indicators and responsive inputs help save hours of repetitive work
LiteFold’s design treats time-to-insight as a core metric.
Removing Long Feedback Cycles
In traditional computational biology workflows, long feedback cycles are one of the most under-acknowledged constraints on scientific progress. Even when state-of-the-art models are available, the time between forming a hypothesis and receiving actionable computational feedback can stretch from days to weeks. Protein structures must be prepared, jobs scheduled on shared compute resources, docking runs executed offline, results parsed manually, and redesigned molecules resubmitted—often across multiple tools and formats. Each iteration introduces latency, friction, and a growing disconnect between scientific intuition and computational output.
What stands out about LiteFold is that it is explicitly designed to collapse these feedback loops and reduce wasted effort. By integrating structure prediction, pocket analysis, molecule generation, and scoring into a single interactive environment, LiteFold transforms what were once batch-oriented, asynchronous workflows into near-real-time exploratory processes. Researchers can modify inputs, regenerate molecules, and immediately observe changes in predicted binding, geometry, or drug-likeness—preserving cognitive continuity between question and answer.
This reduction in feedback latency has second-order effects that are often more important than raw speed. Short feedback cycles encourage exploration over optimization, allowing scientists to test more hypotheses, discard weak directions earlier, and develop stronger intuition about structure–function relationships. In contrast, long feedback cycles tend to push teams toward overplanning and premature convergence, simply because iteration is expensive.
The platform minimizes context switching, removes infrastructure delays, and surfaces results in a way that aligns with how researchers think and reason. In doing so, LiteFold shifts computational biology from a slow, queue-driven process to an interactive design discipline, where insight compounds through rapid iteration rather than delayed computation.
Cloud GPUs Without the Infrastructure Burden
One of the biggest challenges researchers face is limited access to GPUs and the complexity of managing them. LiteFold’s cloud-native architecture removes one of the most significant bottlenecks in modern bioinformatics: GPU availability and setup. Instead of relying on local clusters, complex CUDA configurations, or long queue times on shared academic infrastructure, LiteFold provides on-demand access to serverless cloud GPUs optimized for protein folding, docking, and generative chemistry workloads.
This approach allows researchers to scale compute only when needed, experiment freely, and iterate faster—without capital expenditure, DevOps overhead, or infrastructure lock-in. For small labs and startups, this effectively converts what was once a fixed cost into a flexible operational capability.
Human-in-the-Loop Discovery
LiteFold is designed to enable collaboration between human intuition and AI systems. Researchers can intervene, modify, and guide molecule generation rather than passively accepting black-box outputs, making discovery more creative, transparent, and controllable.
By compressing traditionally sequential discovery pipelines into tight, interactive feedback loops, LiteFold enables workflows that once took weeks—structure preparation, docking runs, molecule redesign, and rescoring—to be completed in minutes or hours within a single environment. This acceleration fundamentally changes how researchers explore chemical space, encouraging broader and earlier exploration in the discovery lifecycle.
Education as a First-Class Feature
LiteFold goes beyond tooling by actively educating its users. Through its blog, explanatory content, and transparent discussion of limitations, LiteFold guides users through its own learning process, helping them understand why models behave the way they do—not just how to use them. This educational philosophy is also reflected in the platform’s UI and its deeply integrated AI research assistants.
This emphasis on education empowers interdisciplinary teams—ML engineers entering biology, chemists exploring AI, and students learning structural biology—to develop intuition alongside technical capability. Over time, this leads to more informed users and better science.
Cost-Efficient Experimentation
By combining cloud compute with AI-guided design, LiteFold enables teams to test more ideas with fewer resources, increasing the probability of discovering viable leads without large upfront investment. Its pay-to-predict model adds scalability, allowing easy expansion without the need to invest in large GPU clusters.
AI-Native, Not AI-Retrofitted
LiteFold is built from the ground up around AI workflows, rather than retrofitting AI into legacy computational chemistry stacks. This results in faster iteration, better user experience, and tighter integration between models and decision-making.
Core Capabilities of LiteFold
Structural Biology Meets AI
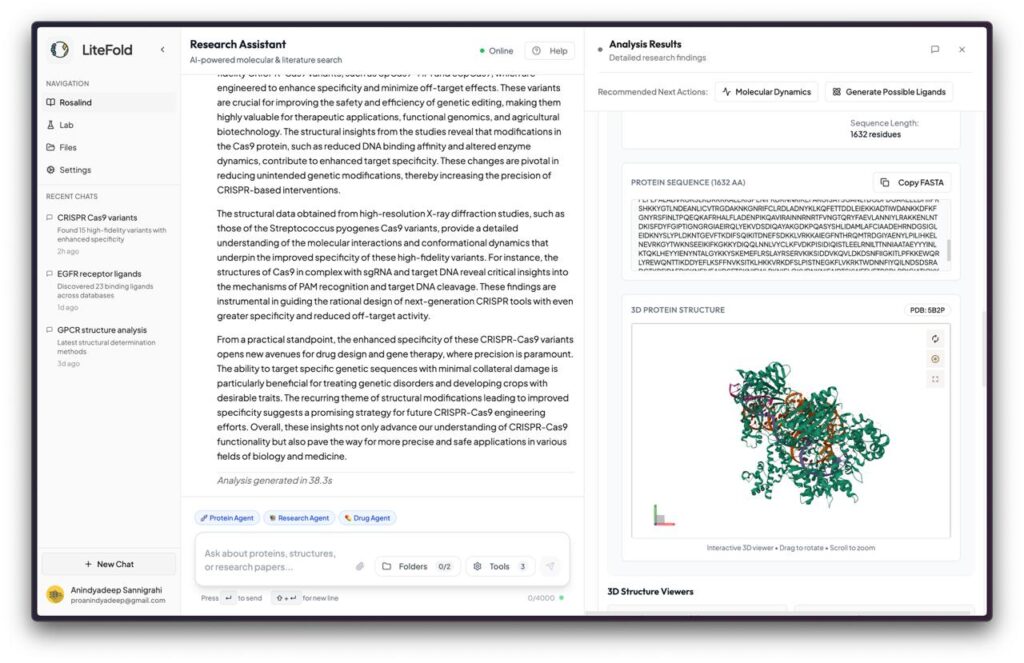
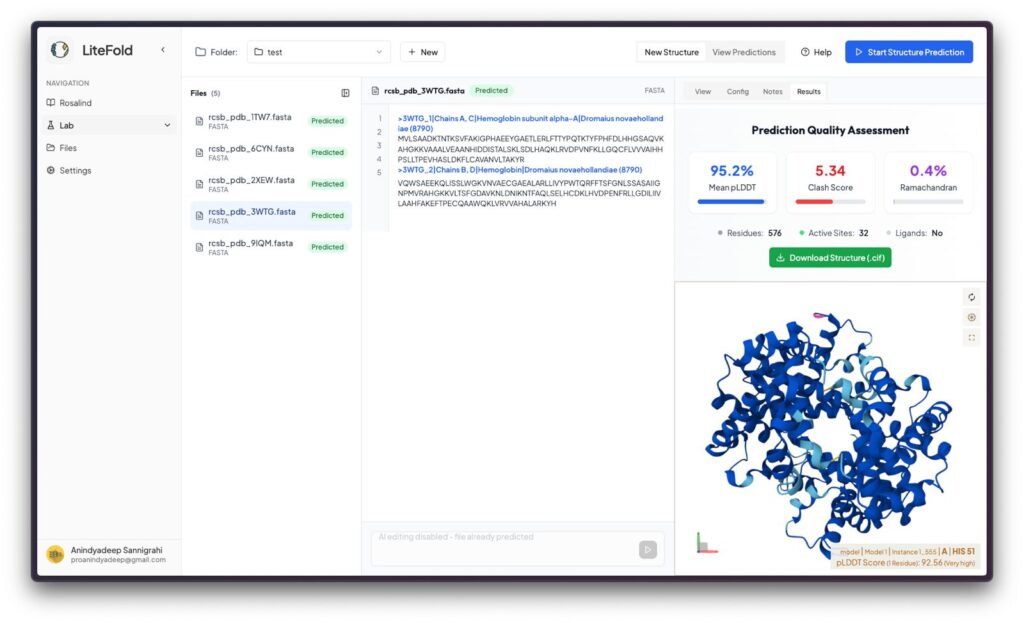
LiteFold’s core tools enable protein structure prediction from sequence—a task that historically required years of experimental work. The advent of deep learning models such as AlphaFold 2, BoltzGen, and ESMFold has revolutionized this space by enabling high-accuracy computational predictions. LiteFold leverages these advances to provide accessible structure prediction and analysis.
The platform allows researchers to:
- Upload sequences and run structure predictions without complex GPU setups
- Visualize predicted 3D structures using interactive tools
- Compare predictions against experimental data and evaluate quality metrics (e.g., pLDDT, TM-score)
This significantly lowers the entry barrier for labs and students without specialized computational infrastructure. Importantly, these models are continuously refined and updated in the background, ensuring users do not need to track rapid developments themselves.
Generative AI in Drug Discovery
Traditional drug discovery has often been compared to searching for a needle in a haystack—experimenting with billions of compounds to find a few viable candidates. LiteFold integrates proven generative models that explore chemical space far more efficiently by learning patterns from existing data.
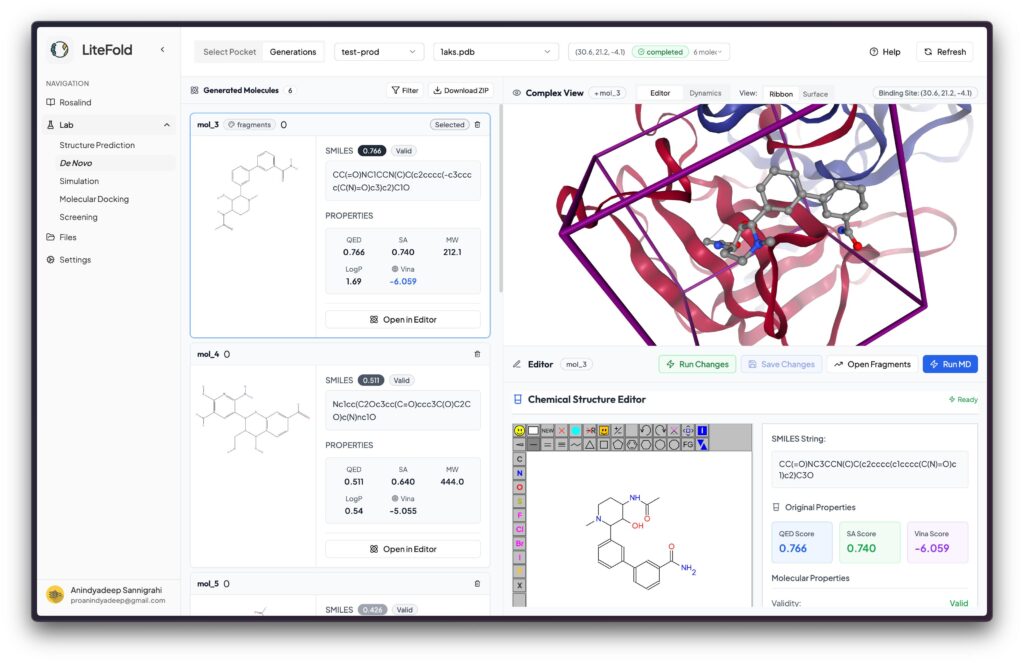
One of LiteFold’s flagship offerings is LiteFold DeNovo, a workflow that enables users to generate novel small molecules tailored to specific protein binding pockets.
Key capabilities include:
- Pocket identification: Upload protein structures or use AI to detect ligand-binding pockets
- Molecule generation: Generate hundreds of novel compounds in real time
- Interactive editing: Refine molecules with instant updates to predicted metrics
- Evaluation metrics: Binding scores (e.g., Vina docking), synthetic accessibility, and drug-likeness (QED)
- Fragment growing: Intelligently expand existing molecules within pocket contexts
- Future dynamics simulation: Planned molecular dynamics simulations to assess binding stability
This workflow condenses weeks or months of traditional computational work into a single intuitive session—representing a paradigm shift toward interactive, AI-guided discovery.
Machine Learning Meets Physics
While fast scoring methods such as molecular docking provide rapid approximations of binding affinity, they often miss important thermodynamic nuances. LiteFold combines physics-based modeling with machine learning corrections to improve predictive accuracy across drug design workflows.
By abstracting complex techniques such as molecular dynamics into user-friendly experiences, LiteFold enables researchers to move beyond static predictions and gain dynamic insights into molecular motion and energetics.
Who LiteFold Is Built For
LiteFold serves a broad audience, including:
- Academic researchers accelerating early-stage discovery
- Biotech startups seeking cost-efficient computational workflows
- Students, DIYers and educators learning structural biology or AI in biology
- Interdisciplinary teams bridging biology, chemistry, and machine learning
Challenges and Future Directions
LiteFold represents a new generation of research infrastructure where AI, physics, and biology converge in a user-centric platform. However, like all AI-driven discovery tools, computational predictions still require experimental validation. While simulations and AI metrics provide powerful guidance, wet-lab confirmation remains the ultimate bottleneck.
Planned future capabilities—including enhanced dynamics simulations, broader screening workflows, and deeper integration of physics-based models—signal LiteFold’s ambition to deliver increasingly realistic and impactful research tools.
As biotech continues to embrace AI and cloud-native research environments, LiteFold stands out as an early example of an integrated, accessible, and interactive discovery platform—bringing advanced in-silico experimentation out of elite labs and into the hands of a much broader scientific community.
Check out Litefold on their website and let us know what you thing about it.