Nanopores Found Useful in Detecting Single Molecule Movements
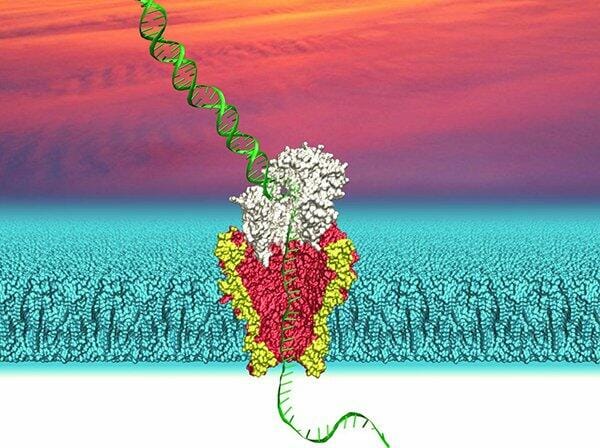
Nanopore’s are proving to be useful for more than just DNA sequencing. The pico sized wonder’s have shown that they can even identify single molecular reactions, such as those between proteins with much better resolution that any other technology has made possible until today.
Unlike the Swiss researchers who used viscous solution to slow down passing DNA. Researchers at Illumina and the University of Washington describe their attempts to improve nanopore for DNA sequencing and stumbling on another property that changed the direction of their research. When studying the passing of a DNA strand through a nanopore, the researchers accidentally came across other signals from the DNA solution. Further analysing this white noise, they realised was the nanopore picking up electrical signals from passing proteins and other molecules. The sensitivity of nanopores has astonished the researchers who began repurposing their research to improve detection of proteins.
While currently the best way to detect molecules is using optical tweezers, which have a resolution of 300 picometers, the single-molecule picometer-resolution nanopore tweezers or SPRNT has a theoretical resolution of 40 picometer. To put this into perspective, a strand of DNA is 1.2 nanometer in width across, allowing close to 900 DNA strands to be inserted at once.
While the resolution of single molecule differentiation is something that future researchers can look forward to, there is another aspect of SPRNT that they will find more astonishing. SPRNT was able to differentiate between mechanisms that cellular proteins use to guide DNA to pass through the nanopore. Thats not all, while the SPRNT technique gave this data, it also showed that these proteins use two sequential chemical processes to push or pull the DNA across. The authors believe this can change the overall outlook of understanding how life works and how to design drugs.
Source: Subangstrom single-molecule measurements of motor proteins using a nanopore doi:10.1038/nbt.3357