MinION Sequences Thousands of Isoforms of the Most Complex Gene Known

Scientists from University of Connecticut have partnered with Oxford Nanopore Technologies to sequence all possible isoforms of the most complex gene known, Down Syndrome cell adhesion molecule 1 (Dscam1), and three other genes, all from Drosophila. The researchers used the MinION nanopore sequencer to obtain 7,899 full-length isoforms. The study has been published in the journal Genome Biology.
Currently, transcripts from a cell are sequenced by high-throughput RNA sequencing. This method is very useful because it allows to obtain the sequence of a gene after splicing, the one conformed only by exons. Tipically, mRNA is primed in its poly-A tail with poly-T oligos, reverse transcribed and amplified. The resulting cDNA is then fragmented and sequenced in small reads that are later assembled by complementarity until the original sequence is reconstructed. The problem comes when dealing with isoforms: different versions of a gene obtained by alternative splicing. Some eukaryotic genes encode from hundreds to thousands of isoforms. With a technique that breaks all isoforms apart in small cDNA pieces it is very difficult to tell which were the original forms or how many of them existed, even using software tools for transcript quantitation. An alternative is Pacific Bioscience sequencing, which can obtain full cDNA lenght reads. However, its high cost makes this option unworkable for most labs. Thus, the team lead by Dr. Graveley decided to try the minION, a relatively cheap sequencing device that generates very long reads.
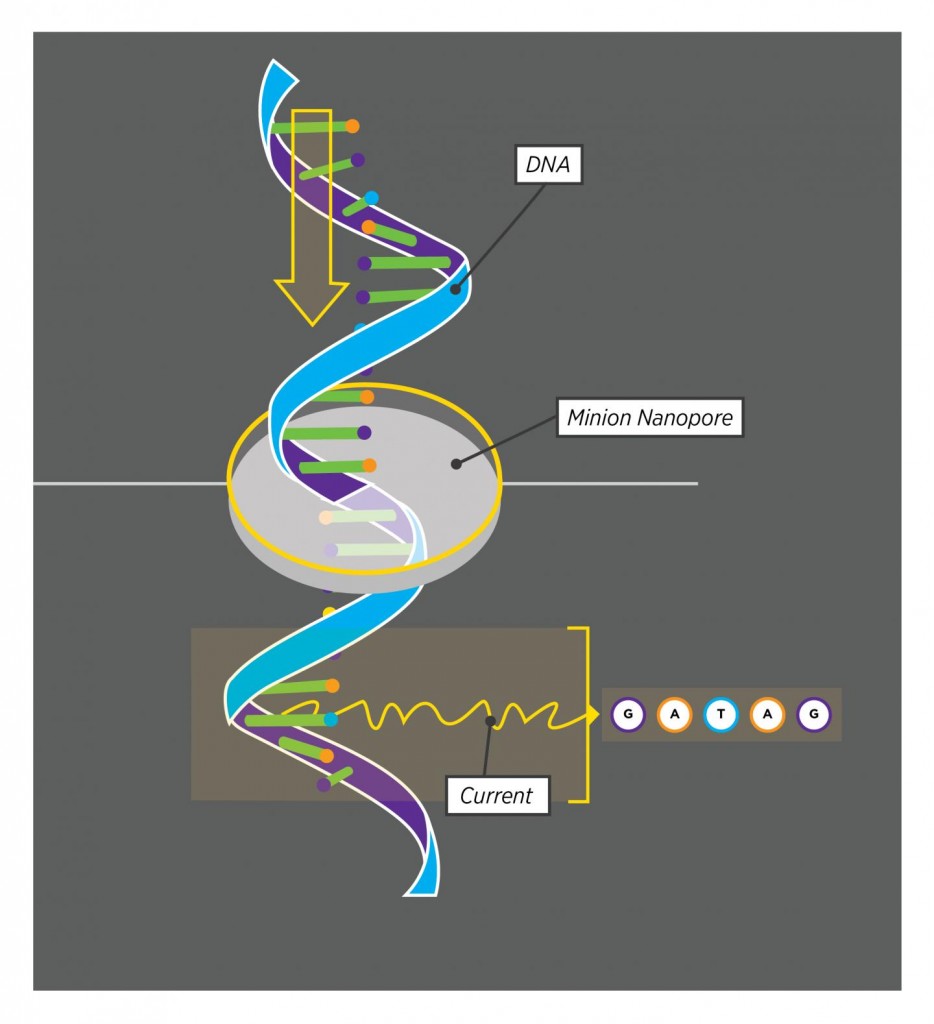
Taking advantage of MinION´s ability to read single molecules
Unlike in traditional methods, the small device reads a single DNA strand that passes through a pore. Five bases are read at a time, and each combination of five generates a specific electrical readout, allowing the MinION to read the sequence without interruption. The UConn researchers analyzed four Drosophila genes, Dscam1 among them. From fruit fly brain cells, they purified and reverse transcribed RNA, and isolated the intended DNAs to pass them through the Nanopore. The team found out that Dscam1 has 115 exons, 95 of them being alternatively spliced, resulting in 38,016 possible isoforms. They sequenced full length cDNAs from all four genes, cataloguing 7899 expressed isoforms.
The successful study will encourage the use of the MinION, a portable and cheap device that is accessible to many labs in monetary and logistics terms.
Source: EurekAlert!