Addgene’s Introduction to the Wide World of CRISPR

![]() Addgene, possibly the largest platform for CRISPR plasmids, libraries, and resources for researchers shines light on this amazing new technology. Here, they give a brief intro on how you can use CRISPR and will direct you to some additional useful resources available at Addgene.
Addgene, possibly the largest platform for CRISPR plasmids, libraries, and resources for researchers shines light on this amazing new technology. Here, they give a brief intro on how you can use CRISPR and will direct you to some additional useful resources available at Addgene.
What is CRISPR?
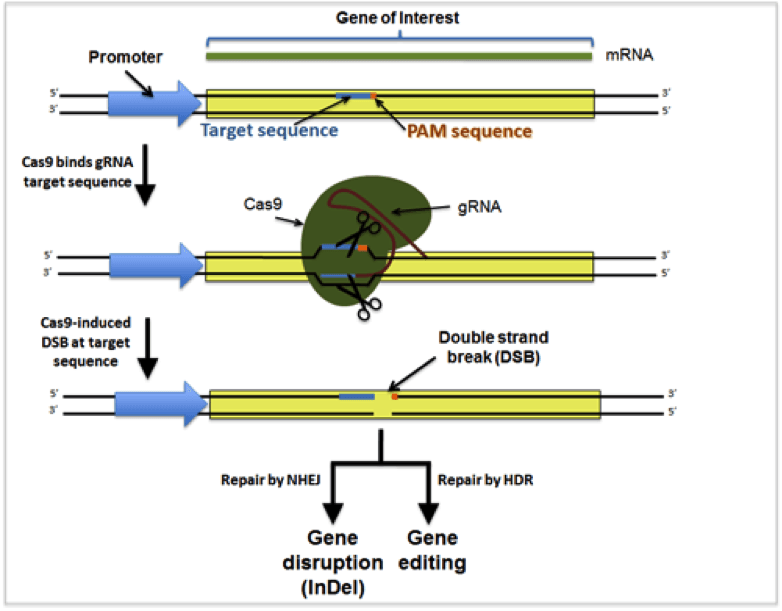
CRISPR stands for Clustered Regularly Interspaced Short Palindromic Repeats. In its natural form, CRISPR is a system bacteria use to defend themselves against foreign DNA (like that found in bacterial viruses), and is commonly referred to as a sort of bacterial immune system. CRISPR systems use nuclease proteins like Cas9 from S. pyogenes to destroy foreign DNA; when foreign DNA enters a bacterium it is inserted into a specific region of the bacterial genome as a form of genetic “memory.” This sequence can be recalled through transcription, and the resulting transcript is used to “guide” a non-specific endonuclease (Cas9) to the foreign DNA for destruction.
There are a variety of types of CRISPR systems, but the one most widely co-opted for genome engineering is the Type II system from S. pyogenes. This system has been greatly simplified such that, for most applications, researchers simply need to express the nuclease Cas9 (or a modified version of it) and a synthetic guide RNA (gRNA) which contains a “targeting” sequence that defines the genomic target for destruction and a “scaffold” region used to bind to Cas9.
How is CRISPR used?
CRISPR is used for 2 main purposes:
- To create loss of function mutations in target genes:
Loss of function (LOF) mutations are generated when the sequence targeted by the gRNA is cut by Cas9 and improperly repaired by the non-homologous end joining (NHEJ) repair pathway (above). Libraries containing many different plasmids expressing many different gRNAs have been generated to allow researchers to screen for a phenotype of interest caused by LOF mutations. These libraries allow researchers to quickly associate a gene or set of genes with a particular phenotype but require next generation sequencing to understand screening results.
- To generate specific mutations in target genomic regions:
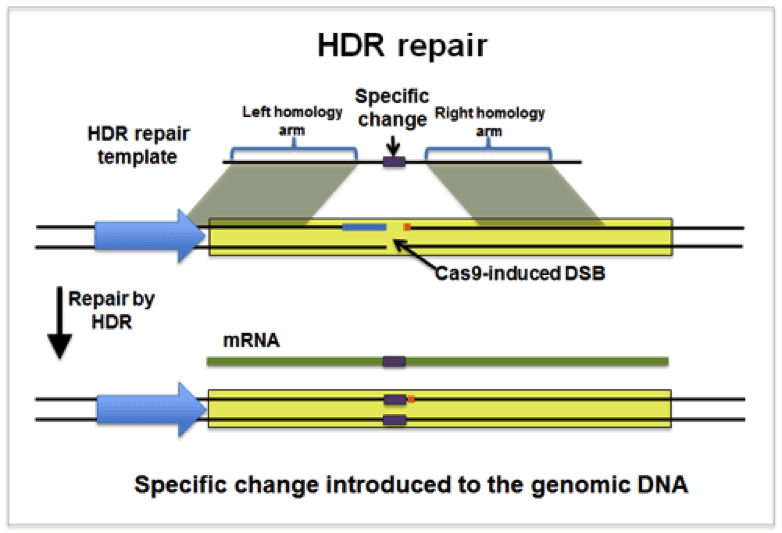
Instead of generating a semi-random LOF in the region targeted by the gRNA, cuts created by Cas9 can be repaired through the homology directed repair (HDR) pathway (above) if a DNA repair template is provided. The researcher designs and co-introduces the DNA repair template along with Cas9 and the gRNA. DNA repair templates have a high degree of homology to the region surrounding the Cas9 target cut site and are incorporated into this site during the repair process. Any alterations contained within the repair template are therefore introduced into the genome when Cas9 cuts are repaired by the HDR pathway. For instance, plasmids that make it easy to add epitope tags onto the genomic copy of a gene have been deposited with Addgene.
When using CRISPR for either of these purposes, off-target effects can be a concern. However, off-target effects can be greatly reduced using versions of Cas9 that must bind to two separate genomic targets to generate double strand breaks (use a nickase or dCas9-FokI for this purpose).
Other uses for CRISPR
- Interfere: Knock down gene expression using “nuclease dead” versions of Cas9 (dCas9) that block transcription of but do not cut targeted regions.
- Activate: Increase gene expression using versions of dCas9 fused to transcriptional activators.
- Purify: Isolate a genomic region of interest using a version of dCas9 fused to an epitope tag – purifying an epitope-tagged, dCas9 under the appropriate conditions will co-purify the target genomic region.
- Visualize: Using dCas9 fused to a fluorescent protein, you can image your target genomic region in live cells.
As you can see, CRISPR opens up many research avenues for scientists who want a quick and easy way to edit specific genomic regions, modify gene expression, and even to purify proteins or genomic regions. CRISPR has been adapted for use in many different organisms. A short list of organisms that can be modified using plasmids at Addgene can be found here.
We hope CRISPR and the resources above will accelerate your experiments and provide you with new research opportunities!
For Additional information on using CRISPR in your research, please see Addgene’s CRISPR Guide and read our CRISPR posts on the Addgene blog.
To find CRISPR plasmids you can use in your research, please see Addgene’s CRISPR pages.
To deposit plasmids with Addgene, please visit our website.
Tyler J. Ford recently received his PhD from the Biological and Biomedical Sciences program at Harvard university and is currently an Outreach Scientist at Addgene. Tyler is particularly interested in Synthetic Biology and Science Communication. Follow Addgene on Twitter @Addgene or follow Tyler directly @tyfordfever.