A New Nanopore Sequencer with Graphene Technology
Researchers from University of Illinois at Urbana-Champaign (UIUC) have developed a new nanopore sequencing method based on graphene nanoribbons that detects double and single stranded DNA in different configurations. The unparalleled thinness of graphene allows high resolution scanning, and electronic sensing of biomolecules is possible thanks to graphene´s semiconductor properties.
There is a growing interest among scientists in finding a cheap, reliable and fast sequencing technique. The most successful sequencer in the last year is the Nanopore, a compact, high-throughput device that promises to advance personalized medicine. In a nanopore sequencing reaction, DNA passes through a nanopore poked on a membrane to which an electrical voltage is applied. When DNA goes through the pore, it causes dips in the electrical current. Reading the magnitude and duration of the electrical drops allows to identify the bases.
Advances in semiconductor nanomaterials allow now to obtain solid, ultra-small structures with electrical sensitivity where pores can be practiced. Semiconductor materials can be doped with acceptor or donor elements, allowing a finer control of the electrical properties. A nanoporous semiconductor layer can be connected to a voltage source, so ion flow can be controlled and the molecules passing through the pore can be sensed.
Nanoporous graphene membranes to sequence DNA
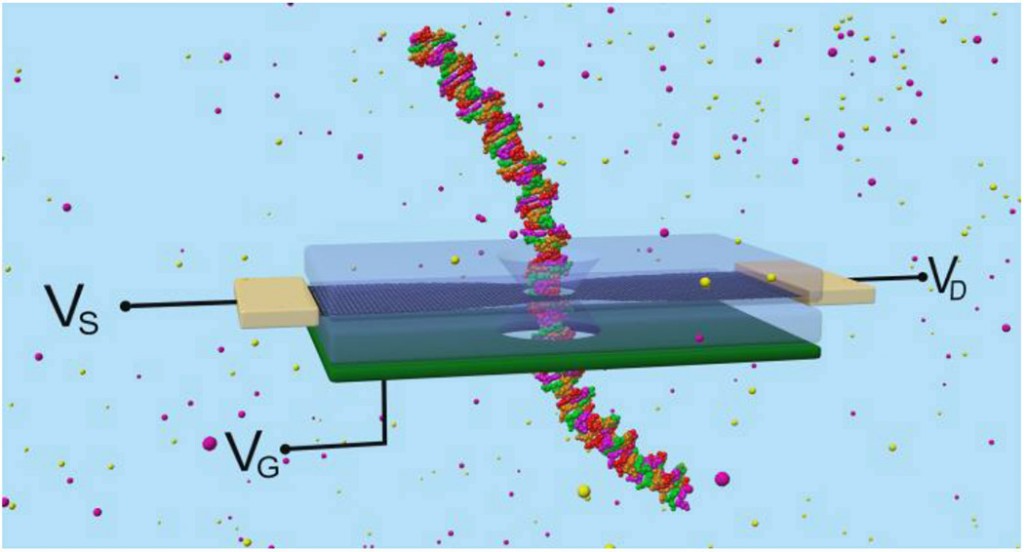
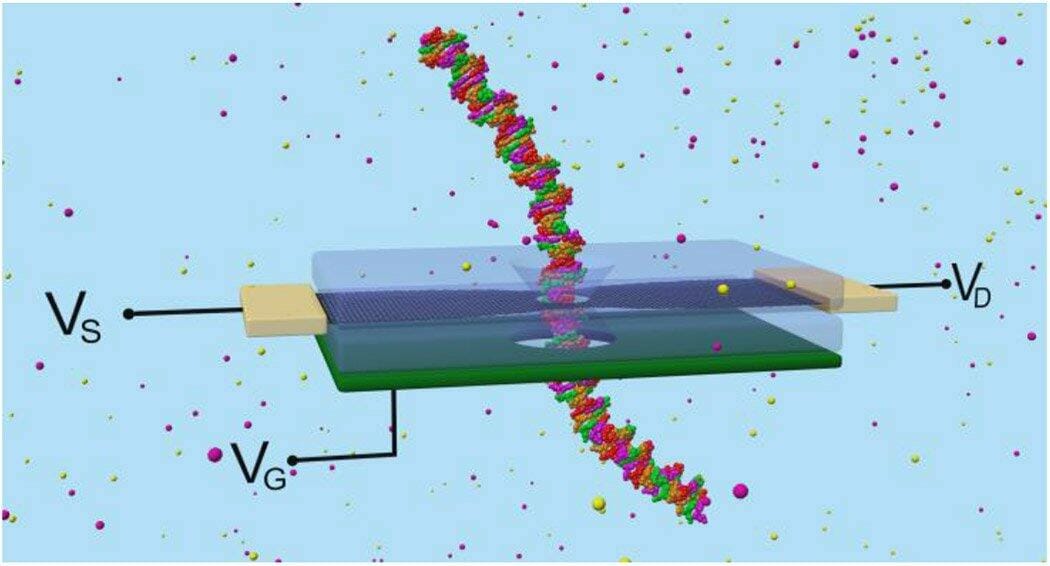
Biolelectronic nanopore sequencer. A silicon layer acts as a back-gate (VG, green). The graphene nanoribbon layer (black) carries the electronic current between the source and drain leads (VS and VD, gold). When a DNA nucleotide passes through the nanopore, the current drops. Sensitivity is enhanced by the bottom gate, which controls the graphene conductance. Source: Jean-Pierre Leburton, University of Illinois at Urbana-Champaign (UIUC).
Based on the latest advances in semiconductor nanomaterials, the researchers developed graphene nanoribbons (GNRs) that could sequence ssDNA and dsDNA in different configurations. The graphene layer one-atom thickness provides unmatched sequencing resolution. Additionally, as a semiconductor material, graphene allows electric control of the biomolecules passing through the pore. A back-gate controls the carrier concentration in the graphene, enabling to optimize sensitivity to the nucleotides. The graphene irregular shape generates many regions of great electronic sensitivity.
The nanodevice was able to detect the helical structure of dsDNA and its helix-to-ladder forced conformational transition. Its high sensitivity -adjustable with changes in carrier concentration or nanopore geometry- allowed to sequence a ssDNA strand.
This graphene-based detector shows great sensitivity and holds promise for developing a portable, high-throughput sequencer that can also detect DNA morphological transformations.
Source: SPIE
