Faster Nanopore Sequencing Simulation Based on Graphene Nanoribbons

Nanopore sequencing technology could be faster and more accurate, according to a simulation by the National Institute of Standards and Technology (NIST). In the recently developed model, DNA passes through a nanohole in the middle of an ultra-thin graphene sheet functionalized with nitrogenous bases that retain its complementary DNA nucleotide, generating a specific electrical signal. The study was published in the journal Nanoscale.
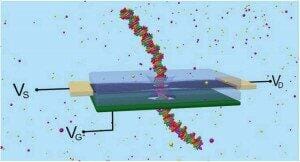
Nanopore sequencing is a recent development of a 20 year old idea by NIST researchers. DNA passes through a nanopore in a membrane and each base is identified according to the electrical changes that the charged molecules produce in the membrane. The technique is very promising: it has been already tested in several laboratories and it has been used to sequence a full genome. Its high speed, reduced footprint and small cost makes it more suitable for field work and point-of-care use than current DNA sequencers. However, there are some accuracy issues that need to be solved, like electrical interference and poor selectivity. To improve the sequencer performance, NIST researchers have developed a model where the nanopore rim would form chemical bonds with the passing DNA, creating a kind of strain sensor that would send different electrical information depending on the difficulty of breaking the bond.
A nanopore with a nitrogenous base-functionalized edge
The researchers conducted simulations of a graphene nanoribbon with a nanopore where several copies of a nitrogenous base have been attached. When the complementary DNA base passes through the hole, they form hydrogen bonds. As the DNA molecule keeps moving, the nanoribbon experiences a tension (deflection) that is detected as a change in electrical current. Finally, the bonds are broken and the membrane goes back to the original position. A full sequencer would contain four stacked nanoribbons, each nanopore attached to one of the four DNA nitrogenous bases.
The new technique could be cheaper and faster than current nanopre sequencing. It is estimated it would be able to process 66 billion bases per second with a 90% accuracy and without false positives.
Source: NIST