3D Map of Human Genome Helps Understand Gene Regulation
Researchers from Whitehead Institute have solved the 3D structure of the human genome in human adult cells and embryonic stem cells (ESCs). Knowing the location of genes and regulatory elements will help to understand gene expression patterns and the influence of nuclear distribution in genetic diseases. The study was published in the journal Cell Stem Cell.
Gene expression is modulated by regulatory elements (trans acting factors) that can be located far away from its target gene, even in a different chromosome, and come in contact with it by DNA looping. Most disease-causing mutations are in fact located in regulatory elements, but many connections between gene and regulator are still unknown. The appearance of this 3D map will be crucial to understand those pathologies.
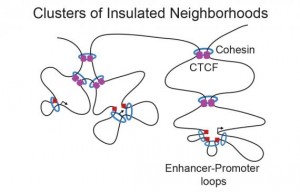
DNA loops are a basic framework for gene regulation
Professor Richard Young had previously done research in genome structure in mouse ES cells. Young’s lab discovered that DNA is organized in loops that compact the chromosomes. Apart from helping to fit very long structures in a small space, each loop creates an independent space that reduces the range of action of a regulatory element to the genes in the vicinity. This study helped to understand the importance of nuclear 3D structure in gene regulation.
In the new study in human cells, the researchers built a 3D map of DNA structure in adult cell and stem cell nuclei. They discovered the existence of 13000 DNA loops tethered by CTCF proteins, and found that each isolated area measures 200 kb and contains one gene. The DNA sequence of the anchored regions is conserved among ES and adult cells, as well as among primates. This means that this organization has been preserved during evolution and is maintained during cell development, thus constituting a basic regulatory and organizational structure. When these structures are lost, the consequences are important: the researchers discovered that many cancer cells had mutations in the CTCF anchors.
Source: Whitehead Institute