Droplet Digital PCR to Measure Tumor Load
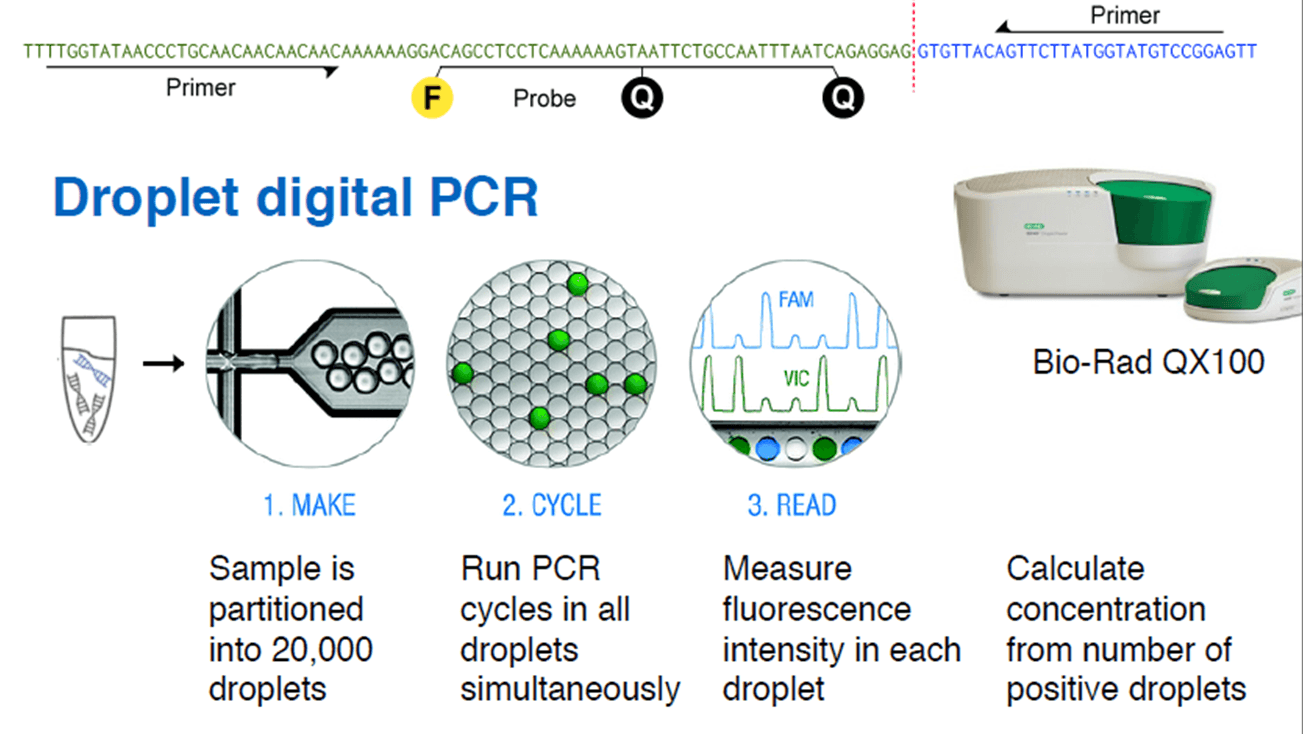
Christof Winter from Technische Universität München presented at Analytica the innovative use of droplet digital PCR to measure tumor load by non-invasive means. The technique takes advantage of the cell-free DNA circulating in blood, which is found in higher concentration in cancer patients.
Cell-free DNA (cfDNA) is found in blood plasma at concentrations of 10–100 ng/ml, in fragments of 150 base pairs. When cfDNA comes from tumor cells, it is called ctDNA, and can be identified by point mutations and chromosome rearrangements typically found in cancer genomes. Translocations, duplications, inversions and deletions leave a tumor-specific fusion region that can be detected and quantified.
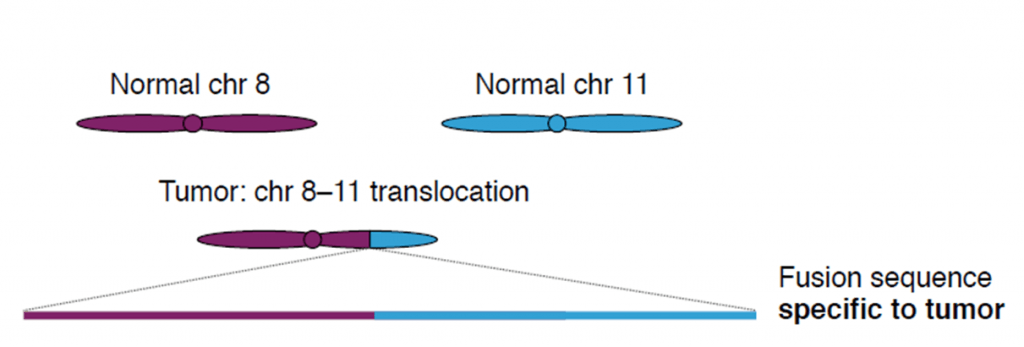
Credit: Christof Winter.
.
First, the whole genomic DNA from a tumor tissue is fragmented into 500 bp pieces. The ends (50 bp) of each fragment are sequenced, and all the read-pairs (100 million) are aligned with a human reference genome, thus identifying the rearranged chromosomes. The exact fusion sequence can be reconstructed with the software SplitSeq, allowing to link chromosomal rearrangements to specific tumors. Finally, the number of rearranged chromosomes can be quantified by droplet digital PCR (ddPCR).
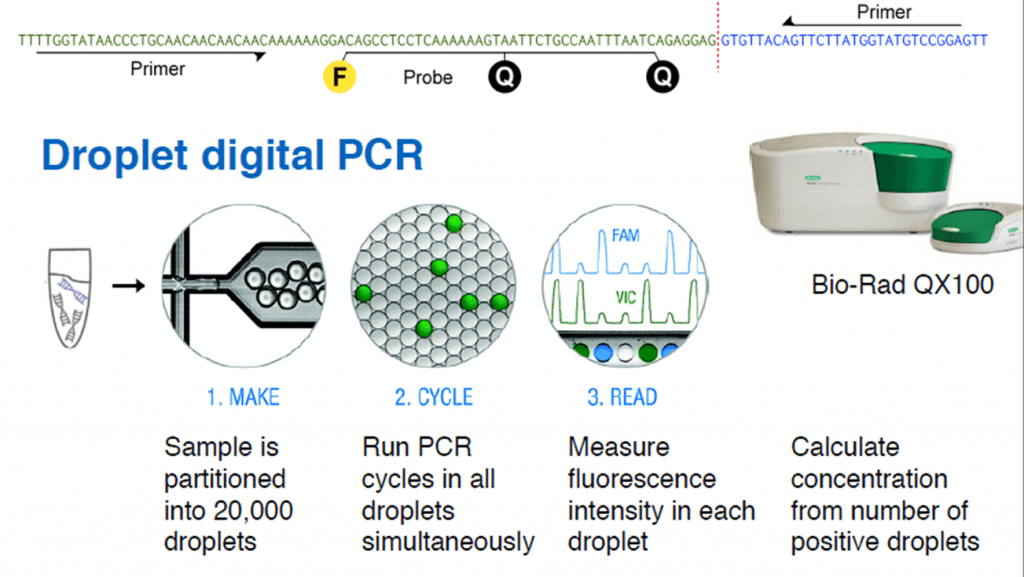
Credit: Christof Winter.
Experiments with a cohort of 20 breast cancer patients have shown that cancer recurrence after surgery can be detected by ctDNA earlier than by symptoms and imaging techniques.
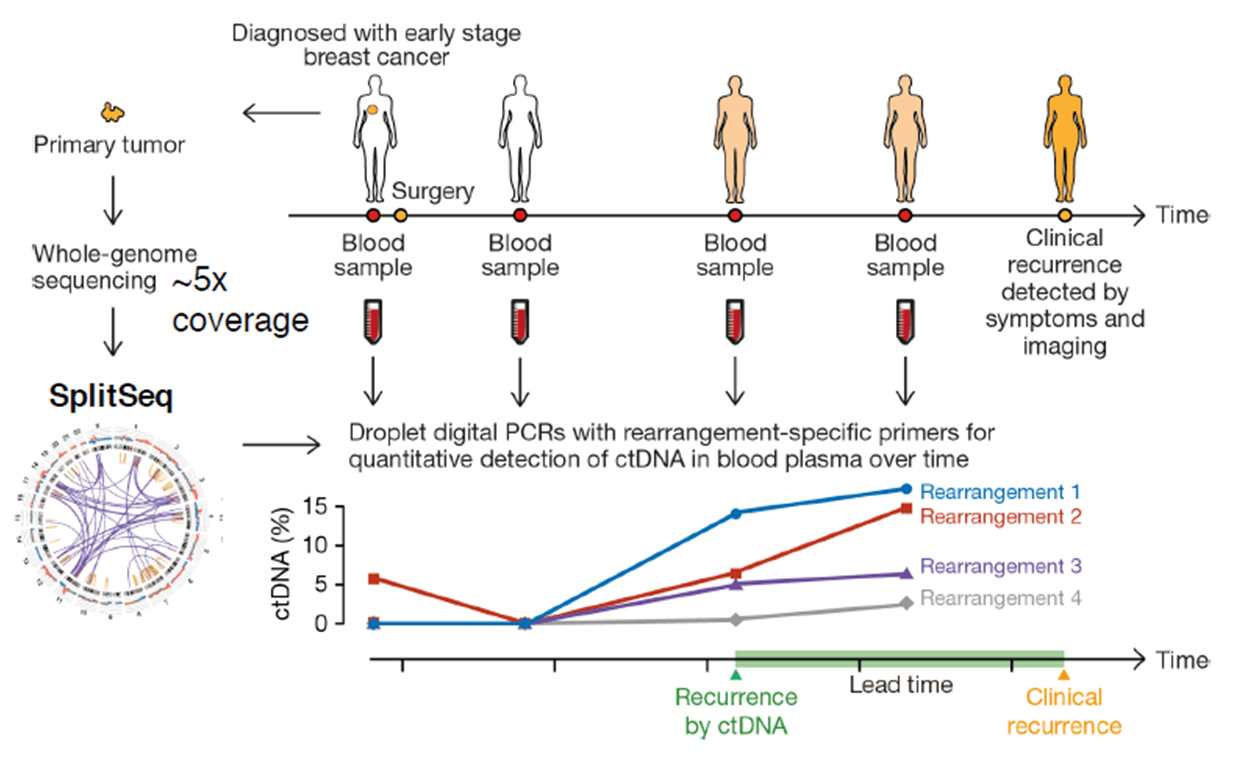
Credit: Christof Winter.
Researchers have also found that ctDNA detection depends on the tumor stage and varies among tumor types.
Currently, ddPCR of ctDNA finds clinical applications like detecting resistance to targeted cancer therapy, monitoring therapy efficacy, detecting relapse early in long-term follow-up and avoiding overtreatment.
You can read more about ddPCR here.

