Quantification of Genome Editing Events Using ddPCR

Bio-Rad presented at Analytica the ultra-sensitive properties of digital droplet PCR (ddPCR) to quantify homology directed repair (HDR) and non-homologous end joining (NHEJ) events in genome editing experiments. Adam Bemis talked about the powerful applications of Bio-Rad’s QX200 Droplet Digital PCR in cutting-edge research and next-gen medicine.
ddPCR divides the sample in thousands of droplets, thus obtaining thousands of discrete measurements instead of one. This allows absolute quantification and single copy resolution, and increases precision and sensitivity. Droplet partitioning increases the relative abundance of rare species.
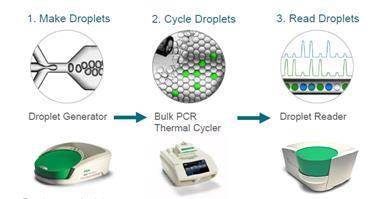
Credit: Adam Bemis.
Genome editing tools are now widely used in molecular biology labs. The CRISPR/Cas system increased efficiency respect to previous editing techniques but, in any case, the endogenous DNA repair systems incorporate changes at low frequency (<5%). Consequently, edited alleles can be hard to detect and quantify.
Gel-based detection methods are insensitive, semi-quantitative and laborious. Clonal/Sanger sequencing is also insensitive. Next generation sequencing (NGS) is costly and time consuming. In comparison, ddPCR offers a sensitive, rapid readout for genome editing events.
ddPCR is sickle cell and HIV research
Several studies published in peer-reviewed journals have already used ddPCR to detect editing events. Bruce Conklin’s group at the Gladstone Institute of Cardiovascular Disease (UCSF) used this technique to detect edited induced pluripotent stem cells (iPSCs) at 100x greater sensitivity than with qPCR.
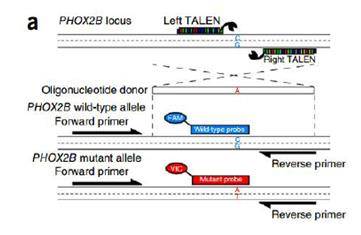
Credit: Adam Bemis.
TALEN was used to introduce a single base mutation in the genome. A fluorescent probe was then added to identify successful edits. ddPCR detected gene editing events with frequency as low as 0.02%. A 10-fold decrease in active work time over existing methods was achieved: the researchers screened 11 clones instead of 2000. Over 20 independent iPSC clones have been isolated to date using ddPCR.
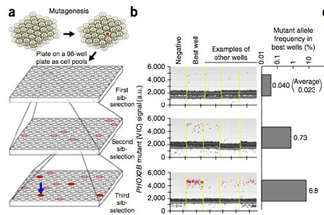
Credit: Adam Bemis.
Conklin’s group has also developed an assay to simultaneously quantify, by ddPCR, HDR and NHEJ events after CRISPR/Cas genome editing. ddPCR has also been used for in vivo therapeutic edit detection and absolute quantification of edited transcripts, to validate the NGS readout in the development of a curative editing strategy for HBB/sickle cell, and for rapid, sensitive detection of NHEJ-induced deletions in HIV.
