A Glowing Reporter for DNA Methylation in Single Cells
Researchers at MIT and Whitehead Institute in Cambridge have designed the first reporter for DNA methylation at the level of individual cells. The method allows to follow the methylation process in real time, both in vitro and in vivo, and to track cell fate changes. The study has been published in the journal Cell.
DNA methylation is the process of adding methyl groups to DNA, which act as molecular tags that switch on and off the expression of the underlying DNA sequence. Usually, methylation will turn off genes, while demethylation will allow their expression. DNA methylation is a central mechanism of epigenetics, the regulatory layer that acts differently on identical genomes, thus changing the fate of the cells that contain them. Because of epigenetics, equal cells of an organism become totally different and build disparate structures like eyes, nails, a heart or a femur. Apart from its importance in development, epigenetics can play a role in disease. Methylation deregulation can have serious consequences, e.g. because of failure to express a tumor supressor gene.
The ability to track methylation in single cells would enormously advance the field. A diseased phenotype could easily be matched with the specific methylation event that did not happen in canonical form, and therapies could be developed. Drugs could be screened for specific gene activation, for example in cancer types due to hypermethylation and gene silencing. So far, however, only snapshots of groups of cells are possible. This method gives a static view of cell methylation, and the information obtained is the result of a mixture of discordant methylation states.
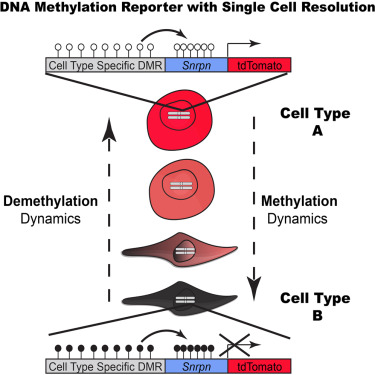
The fluorescent reporter of methylation is inserted near the test locus and is affected in the same way
Postdoctoral researcher Yonatan Stelzer and graduate student Chikdu Shivalila worked directly on this project. Dr. Stelzer conceived the reporter, which consists of a promoter and a gene that encodes a fluorescent protein. The construct can be inserted in CpG islands, which abound in gene regulatory sequences. The reporter of genomic methylation (RGM) reflects what is happening in its vicinity. When the adjacent DNA sequence is methylated, the inserted promoter (Snrpn) is also methylated, and the reporter gene (tdTomato) is not expressed. When DNA is unmethylated, so is the promoter, which is then expressed and allows expression of the glowing protein encoded in the reporter.
CREDIT: Cell.
The authors are excited about the possible applications of their technique. Many questions remain unanswered in epigenetics, and RGM can be extremely useful to unravel methylation and expression patterns. Dr. Jaenisch’s team has already used RGM to follow the methylation dynamics on non-coding regulatory regions, on ESCs differentiation and on somatic cell reprogramming.
Source: Whitehead Institute