Detection of genomic lesions using an artificial DNA base pair

Researchers from the University of Utah have developed a new technique to localize genomic damage sites. The method uses an artificial DNA base pair previously developed at the Scripps Research Institute, and combines it with several other techniques like base excision repair, amplification and nanopore sequencing. The study has been published in the journal Nature Communications.
Genome lesions like oxidation, deamination or depurination can have negative effects on health. The DNA mutations resulting from those injuries are found in several cancer types and in stress-related diseases. The original lesion is often unkown, but based on the mutation profile, researchers deduct there was a problem in the site. A better knowledge of the location and identity of these modifications would help to understand the cause of those mutations. Unfortunately, DNA modifications are relatively non-abundant, and these sites frequently have mismatched base pairs and are pause sites for polymerases, making them almost impossible to amplify by PCR. Several methods have been developed to sequence epigenetic modifications, like single-molecule real-time sequencing (SMRT) and protein nanopores, but they fail to work with real samples derived from tissues, due to the low abundance of modifications. Moreover, DNA lesions like oxidation are more diverse than epigenetic modifications, which adds up to the difficulty of identifying them.
Four steps based on different techniques
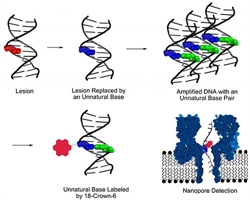
The authors decided to develop a method that would mark the lesions in the DNA, copy and sequence them. First, a base-excision repair mechanism like the one naturally happening inside cells cuts out the lesion. Posterior insertion of the artificial nucleotides dNaM and d5SICS at the lesion site serves to label the region. Without the lesion, the newly introduced base pairs can be easily amplified by PCR. All the copies resulting from the amplification are subsequently labeled with 18-crown-6 ether on the artificial base pair. Lastly, the DNA is sequenced with a nanopore, and the damaged sites can be identified.
The method has proven to be very powerful, with more than ten lesions identified in a single DNA strand. Being able to identify the chemical reactions that lead to DNA mutations will help to prevent cancer and other diseases resulting from those nucleotide changes.
Source: University of Utah