First Ever Direct Image of DNA Structure
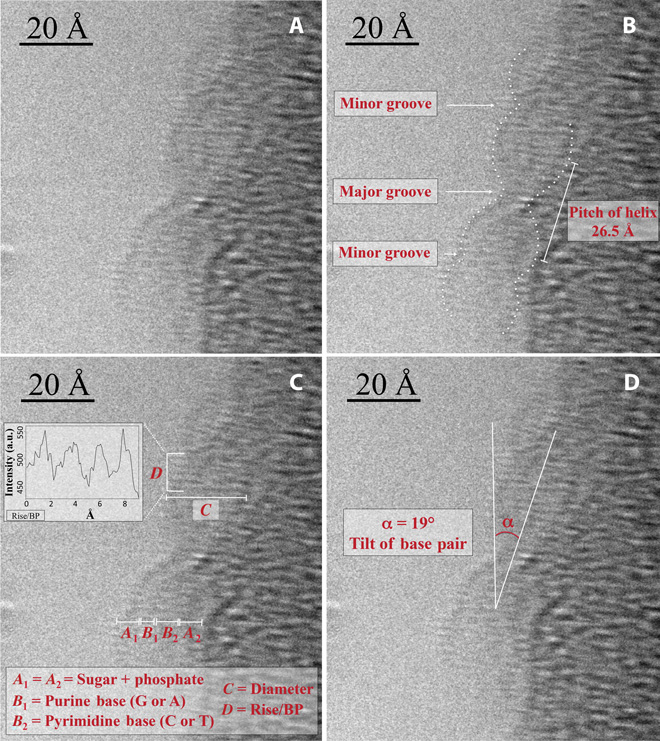
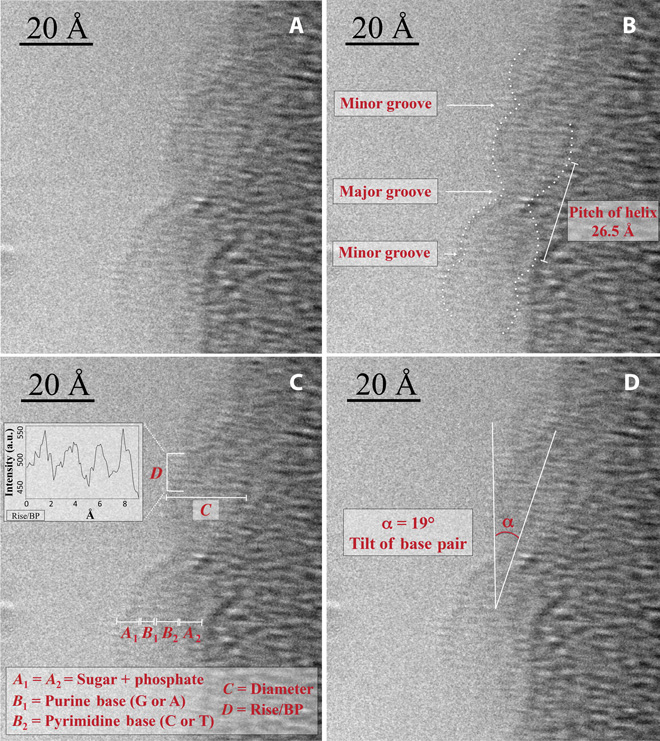
Direct observation and characterization of DNA: double helix, major and minor grooves, pitch of helix, DNA components, tilt of base pairs. From: http://advances.sciencemag.org/content/1/7/e1500734.full
Researchers at King Abdullah University of Science and Technology (KAUST) in Saudi Arabia have developed a technique that allowed them to visualize directly the DNA helix and its inner structure. The team followed a special sample preparation procedure and analyzed DNA by high-resolution transmission electron microscopy (HRTEM). The finding, published in the journal Science Advances, will be crucial to understand biological phenomena at single molecule level.
DNA structure was discovered in 1953 by Watson and Crick, based on information obtained from Rosalind Franklin´s research on the same subject. They identified the double helix conformation in an X-ray diffraction image -the famous “Photo 51”- taken by Franklin and Gosling. Since then, many attemps have been made to obtain a direct image of DNA, for several reasons. An image in the direct space allows to directly see the structure of a single molecule, whereas X-ray diffraction and other indirect techniques obtain average information from a set of molecules. Moreover, X-ray diffraction structures are obtained from crystallized molecules, which is not a physiologically relevant state. Crystallization is also a bottleneck in X-ray diffraction: non-crystallizable molecules are mostly solved by Nuclear Magnetic Resonance (NMR) imaging, but NMR has serious molecular size constraints.
High-resolution transmission electron microscopy combined with an innovative sample preparation method
The KAUST research team wanted to solved a single molecule DNA structure to overcome the problems of indirect imaging and to answer many questions. They wanted to test if direct imaging allows for quantitative information on the characteristic lenghts of DNA, to study local modifications, and to analyze protein-DNA interactions and structure. Direct imaging of molecules is difficult due to the intrinsic low contrast of atoms and the low density of the samples, and there is also the challenge of keeping samples in physiological conditions. This is a problem when using HRTEM that usually compels researchers to increase the electron dose range, thus improving signal-to-noise ratio, but at the price of damaging the sample. Dr. Di Fabricio and his team solved the dilemma embedding the sample in a superhydrophobic microstructured substrate. This sample preparation method proved to be reproducible and favored higher sample purity and density and, crucially, allowed single double stranded DNA (dsDNA) fiber suspension. Thanks to the innovative preparation method, Di Fabricio and his colleagues obtained a direct image of DNA at 1,5 Å resolution, an unprecedented feat. They directly saw the sugar-phosphate backbone, the inner base pairs and hydrogen bonds, and could measure molecular lenghts and geometric features.
This technique, by peeking into single molecule structures in physiological conditions, opens the door to finding the structural basis of diseases, biological mechanisms and epigenetic phenomena, very affected by local modifications. The method will surely be used in the future to solve protein structures, maybe taking advantage of DNA as a scaffold to manipulate the proteins.
Source: KAUST
