Genomic Imprinting Can Target Just a Few Cells
Scientists from the University of Utah School of Medicine have discovered that genomic imprinting can be targeted to subregions of tissues or small clusters of cells. The subtlety of this non-canonical imprinting can affect specific behaviours or moods. The finding has been published in Cell Reports.
Genomic imprinting is an epigenetic mechanism that silences one of the two alleles inherited (from the mother and the father). The silenced copy is said to be imprinted. Imprinting can be very important in insects, where whole chromosomes are silenced for sex determination. Imprinting also determines correct embryonic growth and development in mammals.
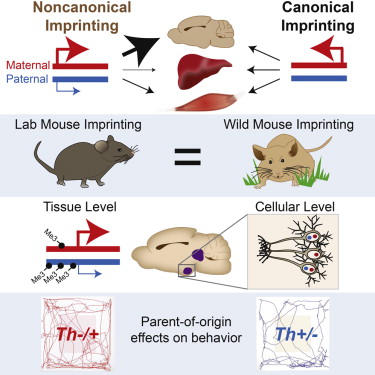
The authors checked genomic imprinting in skeletal muscle, liver and brain regions of live mice. Genomic expression was quantified by in situ hybridization for nascent RNAs. The researchers found imprinted genes in specific tissues that also had different expression in subpopulations of the brain. Specifically, the imprinted neurons belonged to the serotonin and dopamine pathway. This pathway, that affects mood and behavior, has five main genes that are controled in a parent-dependent manner.
Of the 210 imprinted genes analyzed, 80% were subject to noncanonical imprinting, and 64% of those genes showed parental bias exclusively in the brain or subregions of the brain, and not in non-neural tissues, liver, or muscle.
While canonically imprinted genes have just one active copy in nearly every cell examined, noncanonically imprinted genes have one active copy in subsets of cells, and two active copies in others.
The team also discovered that noncanonical imprinting effects are conserved in wild populations.
source: genengnews