Nanopore Technology Can Identify Pathogen Types
Reseachers from Technion–Israel Institute of Technology and Boston University have developed a nanopore that identifies pathogens from their genomic characteristics. The instrument senses single molecules and detects in them large and small deletions or insertions and single nucleotide variations. The study has been published in the journal PLOS ONE.
Fast and accurate identification of pathogens is critical for patients in hospitals and other clinical environments. Incorrect identification often translates into wrong treatments, toxicity, antibiotic resistance, virulence and death. Common molecular identification protocols rely on electrophoresis, where genomic fragments are identified that distinguish benign from harmful microbe strains. However, these procedures are slow, costly, and they are predictably not going to improve substantially in the future. By contrast, solid-state nanopores are relatively new devices that can quickly characterize biopolymers and do not require extensive reagent or sample quantities. Professor Amit Meller and his team decided to adapt the nanopore technology to the detection of genomic traits that differenciate microbiological strains.
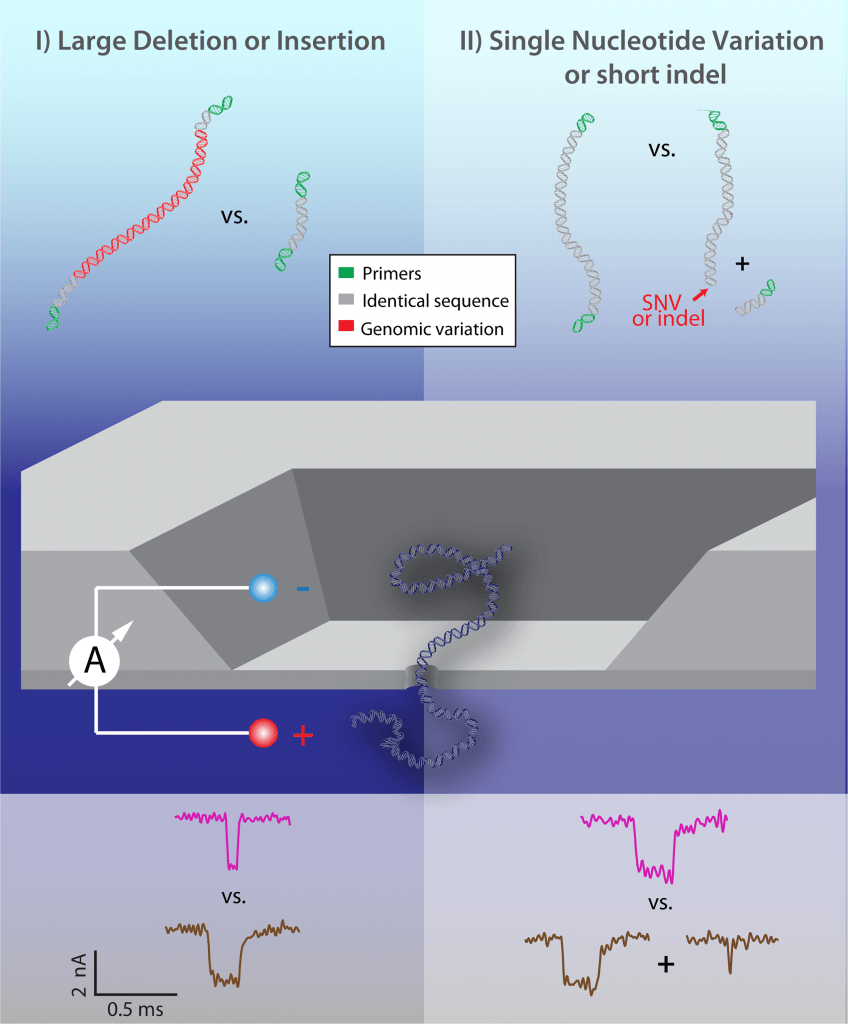
Instead of reading the whole genomic sequence of a microorganism, Amit and his colleagues hypothesized that it would be enough to identify just a particular characteristic that is different between strains already sequenced. This is a simpler question, faster and easier to answer. It could be, for example, identifying a virulence gene, or a drug-resistance gene. The Technion team built a nanopore device from silicon that could identify these differences by detecting two types of variations.
Two reading modes: DNA fragment lenght and nucleotide variation
In the first reading mode, the nanopore device can detect the lenght of a DNA molecule. When measuring the electrical current in the nanopore, it is observed that DNA causes a current blockage when passing through the pore. Based on the DNA translocation time, as reflected in the electrical readout, the researchers can infer the presence of an insertion or deletion.
In the second mode, DNA is previously digested with a restriction enzyme known to cut a specific nucleotide sequence, which might be present only in the virulence or resistance gene that characterizes a pathogenic strain. If the nanopore senses only one molecule, the measured current will consist of only one current dip. If the gene is present, however, the DNA will be cut into two molecules, and then two dips will be observed.
Credit: Technion.
The same method is used in nanopore sequencing, but measuring current blockade and dwell time in greater detail, so that it allows to distinguish nucleotides. The new system was tested on Mycobacterium tuberculosis and Streptococcus aureus, and the authors concluded that confident results were obtained in less than a minute.
The team of researchers is currently working on a device that could detect multiple variations at the same time, with several nanopores running on parallel and microfluidic chips for multiple enzymatic digestions. They expect to have a commercial version of their nanopore device in a few years.
Source: genomeweb