Six New Tools for the Study of Protein, DNA and RNA Motifs
Researchers from the University of Queensland, Brisbane and the University of Washington, Seattle launched in 2009 the MEME Suite, a web-based service for the study of protein, DNA and RNA motifs. Six new functionalities have been added to the site since then, so the creators of MEME Suite published in Nucleic Acids Research a paper that explains the new tools capabilities, gives advice on how to use them and presents some practical examples.
Sequence motifs are short DNA, RNA or amino acid patterns conserved through evolution. They usually indicate a site of biological relevance, like an active protein site or a DNA binding domain. The identification of these motifs is crucial for understanding the molecular mechanisms underlying cellular processes and diseases. In this scenario, it´s easy to see why MEME Suite has been used in more than 9800 published studies and had 20000 unique users in 2014. Its use is predicted to grow in the current age of big data, genomics and proteomics.
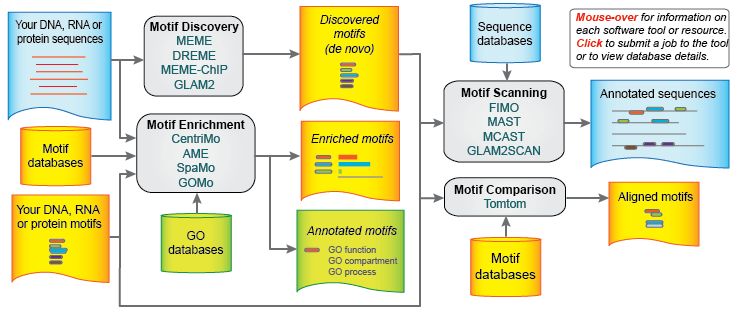
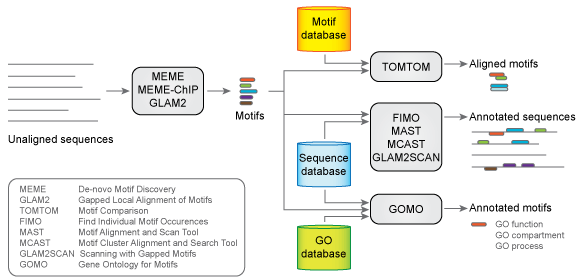
The web-based version of the MEME Suite includes 13 tools for performing motif discovery, motif enrichment analysis, motif scanning and motif–motif comparisons. The core of the suite is the MEME motif discovery algorithm, which finds de novo motifs in the user-provided sequences. The MEME Suite provides a large number of proteomic and genomic sequence databases for motif scanning and many motif databases for motif comparison.
The last six tools added to the suite
Six of these tools—DREME, MEME-ChIP, CentriMo, AME, SpaMo and MCAST had not been presented in the last paper about the MEME Suite. DREME is used for motif discovery in transcription factor ChIP-seq data. MEME-ChIP is used for motif analysis of large DNA datasets. CentriMo infers direct DNA binding from ChIP-seq. AME determines DNA-binding transcription factors in a given set of genes by detecting enrichment of known binding motifs in the genes’ regulatory regions. SpaMO infers transcription factor complexes from ChIP-seq data. MCAST searches for statistically significant regulatory modules in genomic DNA.
The site is available for free for academics at http://meme-suite.org.