3D Protein Microcrystal Structure Identification by EM
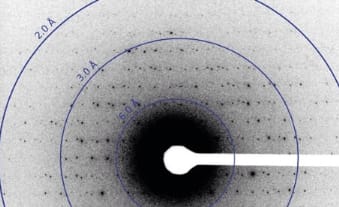
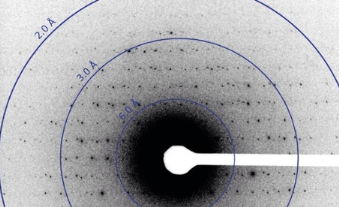
Image shows electron diffraction from lysozyme microcrystals prepared by Tamir Gonen’s laboratory at HHMI’s Janelia Farm Research Campus. Image: Courtesy of Gonen Lab
Researchers at Janelia Farm Research Campus at HHMI are reporting on a new technique of imaging 3D protein microcrystals using electrons from an electron microscope. The size of crystals required for the new technique is a million times smaller than that required for commonly used X-ray crystallographic studies.
Currently researchers spend years trying to get the perfect protein crystals in large sizes to form. It is often met with unusable crystals, very small crystals or the protein fails to crystallizes. Researchers at Janelia led by Dr. Tamir Gonen developed the new technique called MicroED (micro electron diffraction) to accelerate protein structure imaging. The researchers used the fact that it is easier to generate small perfect crystals rather than large crystals. By using electron microscopes the researchers we able to analyze 3D crystals.
Electron microscopes have been used on 2D crystal analysis before, however they were not used on 3D crystals because the radiation from high energy electron beam destroys the 3D crystals before researchers can record enough diffraction patterns. The crystals can only handle two or three low dose exposures, which leads to incomplete data – making painting a clear picture of the complete structure tough. These challenges become even more insurmountable when applying the same 2D method to 3D crystals. There is no way to find out what patters represent in the reciprocal space of the crystal.
The Janelia researchers reduced the dosage to 200 times less than what is considered a low dose to preserve the crystal structure, which made possible collection of multiple data sets out of a single crystal, which would have otherwise been impossible using the conventional low dose radiation beam. The researchers were able to collect 90 data sets from a single miniature crystal of lysozyme, each at 1 degree divergence. Using custom software the researchers were able to stitch together the diffraction patters to obtain the complete structure of lysozyme. The researchers achieved a resolution of 2.9 angstroms in their experiment, providing enough differentiation of the protein’s different structures. They expect to achieve a resolution of 1.7 angstron by further tuning the technique and the software.
Paper: Three-dimensional electron crystallography of protein microcrystals (DOI: http://dx.doi.org/10.7554/eLife.01345)
