Accurate and Simple Elastic Model of RNA Dynamics
Researchers from the Scuola Internazionale Superiore di Studi Avanzati (SISSA) in Trieste, Italy have found a simple but accurate method to study RNA dynamics. In the study, published in Nucleic Acids Research, Bussi and colleagues show that an elastic network model (ENM) of 3 beads per nucleotide has the right balance of accuracy and complexity, as validated by molecular dynamics (MD) and biochemical experiments.
MD is a computer simulation of molecules and atoms movements. All particle interactions are represented as interatomic potentials or molecular mechanics force fields. This method is the most accurate for representing the dynamics of biomolecules. But it is also time- and computer-consuming, with long-week calculations.
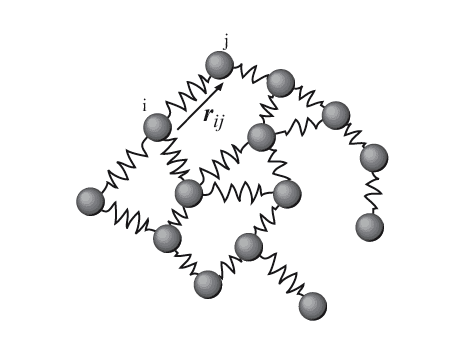
ENM is a cheaper alternative to MD, and has been used to study protein dynamics. In this method, atoms are grouped in beads, that are connected by springs. A high (or low) number of atoms per bead will give a coarse (or fine) simulation. Researchers try to find the best balance between a faithful representation of molecular dynamics and a reasonable computer consumption. Researchers are becoming more interested in RNA dynamics, as evidence points to the importance of RNA movements in its function.
3-beads-per-nucleotide ENM can simulate RNA dynamics with accuracy
A team from SISSA has been using ENM to study RNA, and they propose that coarse simulations are still faithful to real RNA dynamics. Giovanni Pinamonti, Sandro Bottaro, Cristian Micheletti and Giovanni Bussi propose using three beads per nucleotide. The three beads represent the sugar, nitrogen base and phosphate. The results were compared with a full MD simulation, with satisfactory results, validating the 3-beads-per-nucleotide ENM as the most balanced between accuracy and resource consumption.
The ENM results were also compared with SHAPE (selective 2′-hydroxyl acylation analyzed by primer extension), a chemical probing experiment that informs on RNA structure at nucleotide level. SHAPE results validated the 3-beads-per-nucleotide ENM predictions too.
Source: genengnews