A More Precise Nanopore Sequencer Based on Polymer-Tagged Nucleotides
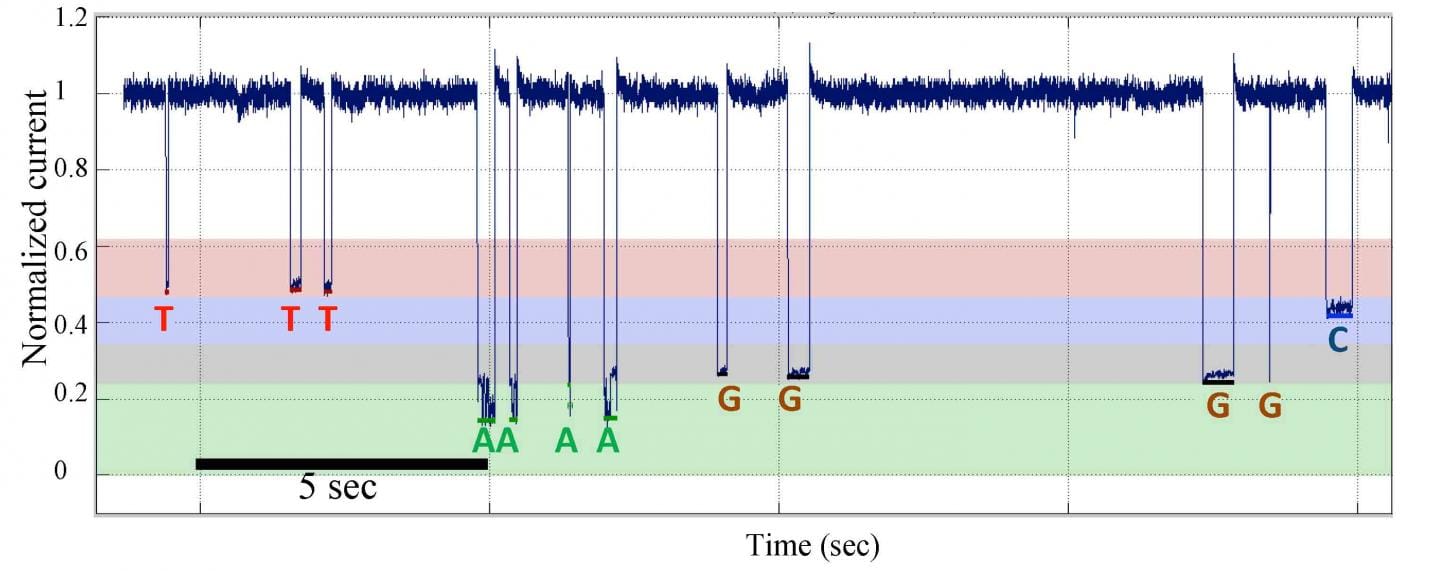
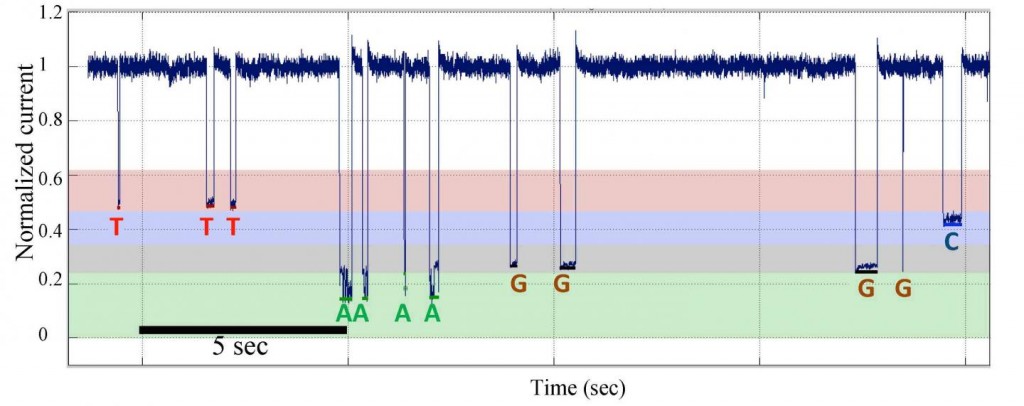
Single-molecule nanopore DNA sequencing by synthesis data from a template with homopolymer sequences. (http://www.pnas.org/content/early/2016/04/14/1601782113.full)
Researchers from Columbia University School of Engineering and Applied Science have developed an advanced real-time, single-molecule nanopore sequencer. In collaboration with Genia Technologies (Roche), Harvard University and the National Institute of Standards and Technology (NIST), the engineers built an electronic DNA sequencer with better resolution than current nanopore devices. The study was published in the journal PNAS.
Genome sequencing has allowed the implementation of personalized medicine, increasing the efficacy of diagnoses and treatments. Classical high-throughput DNA sequencing methods rely on optical detection of the four nucleotides, but alternative technologies are being explored to make sequencing cheaper, faster and more accurate. Nanopore sequencers, the most recent achievement in the field, are miniaturized devices that identify single nucleotides from a strand of DNA passing through a protein channel. An electrical voltage is applied to the pore, and each DNA base produces a specific electrical signal that is detected, thus allowing to read the sequence. However, because of the similar chemical structure of the four nucleotides, the method can’t accurately distinguish them.
Tagging each of the four nucleotides with a different polymer
The engineers from Columbia University conceived a new sequencing method based on a principle they reported on an earlier study: sequencing by synthesis (SBS). Like in orthodox nanopore sequencing, the system relies on electronic detection of single molecules passing through a protein channel. But there are several innovations in the nanopore SBS system, which is described in the PNAS paper. The new nanopore array consists of a protein channel covalently attached to a polymerase that is bound to a primed DNA template. Additionally, they modified nucleotides by attaching four different polymers to their 5′-phosphate -one for each base type. When the tagged nucleotides are added to the array, the polymerase incorporates the bases for DNA replication, and the tags are captured in the pore, where they produce a particular electrical signal. The polymer tags signals are much more different among themselves than those of the natural nucleotides, increasing the accuracy of the sequencing method.
The researchers have observed higher throughput and performance that they predicted, and read lengths over 1000 bases are already feasible. Future improvements in the SBS sequencer will enable its use in routine diagnoses.
Source: Columbia Engineering
