A New Software to Improve CRISPR/Cas9 Targeting
Scientists from Harvard and University of California have designed an online tool to optimize CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 targeting. The researchers compared genomic DNA (gDNA) and CRISPR guide RNA libraries for more than 1400 gene loci. The library-on-library method allowed to obtained common patterns for the most active and specific guide RNAs. The study has been published in Nature Methods.
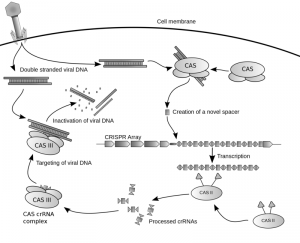
Some prokaryotes present the CRISPR/Cas immune system. When foreign DNA enters the cell (usually from a virus or a bacterial phage) it is recognized by the system and incorporated as a spacer between the CRISPR loci repeats. After transcription, RNA from the CRISPR loci is processed into crRNA, a guide RNA that binds Cas endonucleases and targets them to cleave alien, complementary DNA sequences. This acquired immunity mechanism was seen by the Doubna lab in Berkeley as a potential genome editing tool. They engineered the crRNA sequence to match that of any desired locus in the genome, which would then be targeted by Cas9 and nicked. The system allows to add and delete nucleotides to specific DNA loci in multicellular eukaryotes, without need for markers. The method has been used already to immunize industrially important bacteria against phages, to mutate several genes in human induced pluripotent stem cells (hiPSCs) and for simultaneous mutation in mouse cells and whole organisms. CRISPR/Cas is a potential treatment for human genetic disorders.
A software that finds the best sgRNA by analyzing the activity of previously designed sgRNAs
But finding the proper guide RNA is time consuming. Each target gene could be cut at many spots, so there are many single guide RNA (sgRNA) design possibilities. The problem is that not all sgRNAs are equally active, so researchers have to try several designs to choose the best one. Church and colleagues decided to carry out a library-on-library study comparing thousands or sgRNAs with thousands of DNA targets. They were able to find the common motifs in the most specific and active sgRNAs, and then apply those principles to design the best possible sgRNA for a given DNA sequence. The tool is available online for free at https://crispr.med.harvard.edu/sgRNAScorer/ and a standalone version of the software can be downloaded too.