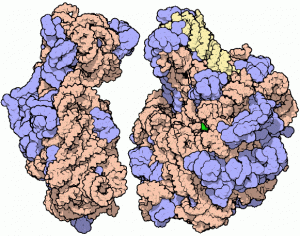
Ribo-T : Ribosome with Linked Subunits Reads Desired RNA
Scientists from the University of Illinois at Chicago and Northwestern University have engineered the Ribo-T, a tethered ribosome that could be used to produce drugs and new materials and help understand how ribosomes function. The breakthrough has been published in Nature.
Ribosomes are ribonucleoproteins, present in all cells, that synthesize proteins. There are two ribosomal subunits: the small subunit which reads the messenger RNA, and the large subunit which joins amino acids to form a polypeptide chain. After the protein is synthesized, the two ribosomal subunits disassemble. The authors found that this was a problem for artificial ribosome design: the natural and synthetic subunits could get mixed and create non-working assemblies. They decided to link the two subunits and created the Ribo-T, a ribosome permanently assembled by a shared ribosomal RNA made of the 16S and 23S rRNAs, which are in the small and large subunit. The Ribo-T was able to synthesize proteins in vitro and in vivo, with capability to keep alive cells devoid of natural ribosomes. The team also designed ribosomes that read only a specific kind of RNA.
The study paves the way for future modifications of the ribosomal catalytic center, so far untouchable. Any wrong mutation in the area causes the ribosome to stop working, and that leads to cell death. The new work by Dr. Mankin and colleagues brings the technology to customize the catalytic center and synthesize polypeptides with new functions. Cells could become factories housing ribosomal machinery designed to produce the desired compounds, like degradation-resistant drugs made of artificial amino acids.
Source: TheScientist