RNA Gets a Database to Call its Own

The RNAcentral Consortium is announcing the launch of RNAcentral a unified resource for non-coding RNA from all species. The open source RNA database has currently 8 million sequences aggregated from expert databases and partner institutes from around the world.
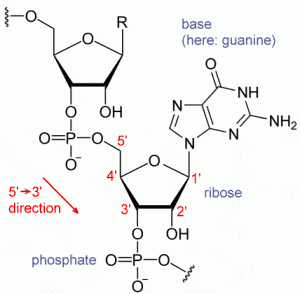
RNA although an important part of the genetic makeup, until now had no database of its own. It was discovery in 1950’s as an intermediate molecule between DNA and proteins. The last decade has seen a huge increase in the number of non-coding RNA gene discoveries, including new classes such as piRNAs, microRNAs and experimental characterization of these RNA components. A large number of discoveries have also pointed out the major role played by RNA in diseases and plants. Despite this, it was difficult for researchers to access RNA database, since key data resources for non-coding RNAs had not yet been created. Unlike proteins, there has been a lack of comprehensive RNA sequence database.
Until now each subtype of RNA had been maintained separately, with researchers using multiple independent resources such as HAVANA for IncRNAs and miRNase for microRNAs to find RNAs encoded by specific genome. RNAcentral brings together data from ten different expert databases and gives each database an accession number for easy reference. It also has faceted search that allow easy RNA search based on species, source and molecular function.
RNAcentral 2.0 will include data from more expert databases, additional data types and information on RNA structure, functions, modifications and molecular interactions.
10 Expert Databases included in RNAcentral 1.0
Check out http://rnacentral.org/ for more information